Recently, Associate Professor Lijie Zhou and his team from the College of Chemical and Environmental Engineering of Shenzhen University published the latest research progress on《Chemical Engineering Journal》(impact factor 15.1, Chinese Academy of Sciences Q1, TOP journal). The title is “Keystone bacteria in a thiosulfate-driven autotrophic denitrification microbial community”. Associate Professor Lijie Zhou is the first author and corresponding author of the paper, and Shenzhen University is the first author unit and corresponding unit.

Recently, some autotrophic denitrification processes have been developed to be alternatives to conventional heterotrophic denitrification processes. Among which, processes using sulfur-based compounds as electron donors seem to have achieved nitrate removal treatment efficiencies comparable to the heterotrophic denitrification processes.
Among all the investigated sulfur components, thiosulfate is often regarded as an excellent electron donor for autotrophic denitrification due to its high water-solubility and non-toxic nature. Although the thiosulfate-based denitrification is an efficient process, the dissimilatory oxidation of thiosulfate needs multiple enzymes, collectively known as the sulfur oxidation (SOX) system, to play in concert to oxidize thiosulfate to sulfate. Although microbial community and functional genes have been measured, complex community and incomplete functional genes analysis led to unknown mechanism of thiosulfate-driven autotrophic denitrification. Consequently, keystone bacteria are needed to clarify in thiosulfate-driven autotrophic denitrification microbial community for treating wastewater with high nitrate concentration.
Five laboratory-scale anoxic bioreactors, designated as S0-S4, were fed with synthetic wastewater containing 120 mg NO3--N/L. Meanwhile, thiosulfate was used as the sole electron donor and provided varying ratios to these bioreactors, resulting in (S/N ratios of 0, 0.7, 2.4, 4.0, and 5.7), respectively. These bioreactors were run in a 104-day period, which was composed of eight operational cycles. The microbial ecology analysis revealed that Proteobacteria (42–66%), Bacteriodetes (15–30%), and Firmicutes (15–28%) were the major phyla residing in the bioreactors. Thiobacillus, Pseudomonas, Arenimonas, and Thauera were identified as predominant genera shared across the bioreactors. Functional genes analysis further showed that only Thiobacillus and Thauera contained the complete nitrate reduction pathway. Due to Thiobacillus was absence of SoxCD for catalysis from SoxYZ-S-SH to SoxYZ-S-SO3, Thiobacillus and Thauera could work collaboratively on the SOX pathway as the keystone bacteria to achieve thiosulfate oxidation.
(a)
(b)
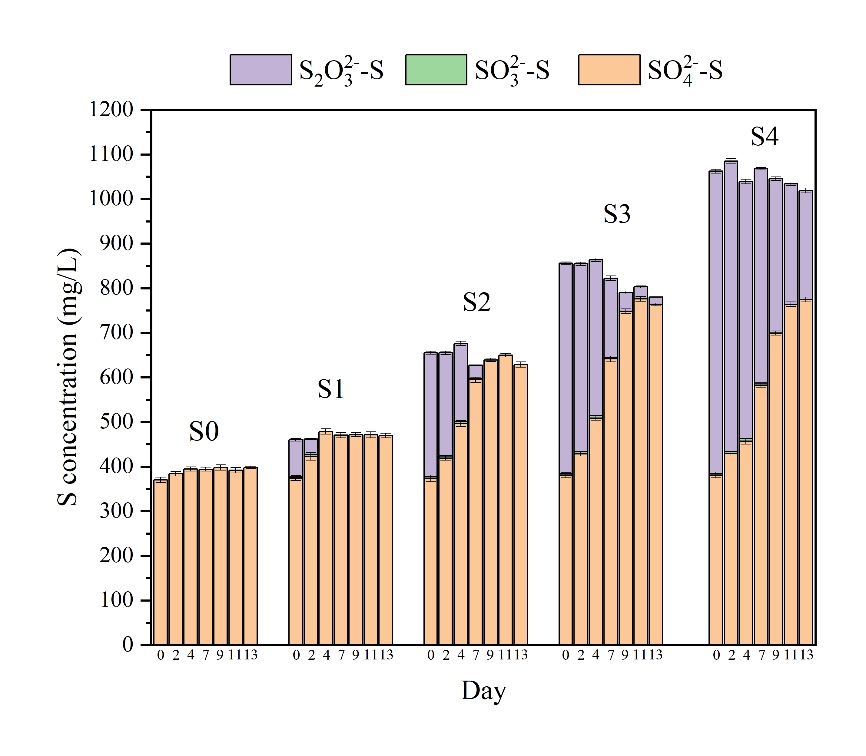
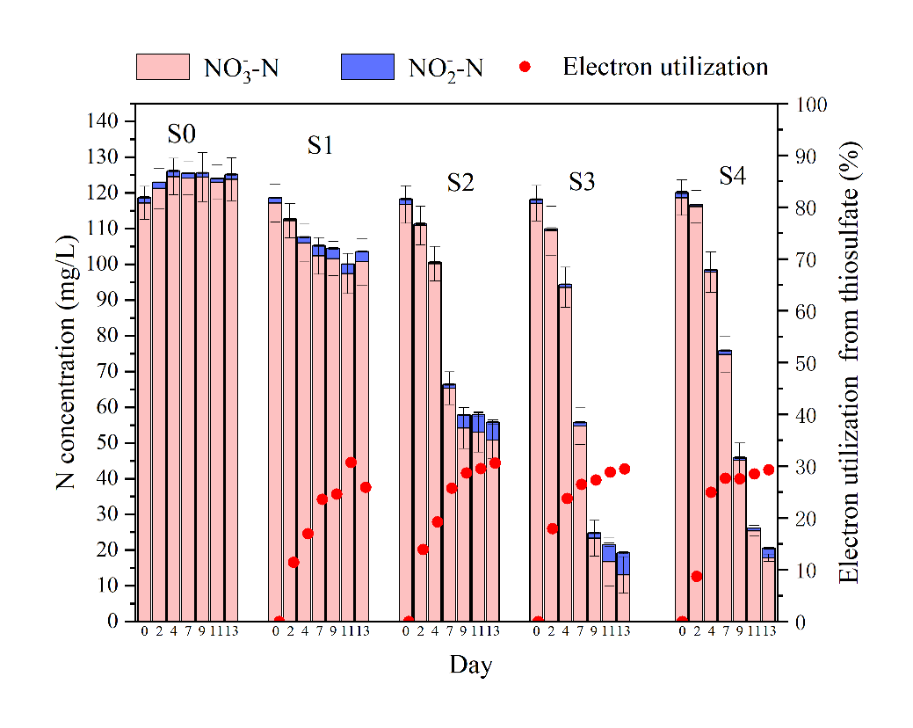
Fig. 1. Variations of (a) sulfate, sulfite, thiosulfate, (b) nitrate, nitrite and electron utilization from thiosulfate during bioreactors operation.
As shown in Fig.1, S1-S4 bioreactors had an obvious nitrate removal with thiosulfate as an electron donor, while S0 bioreactor hardly had any nitrate reduction. Moreover, S1-S4 bioreactors had, respectively, 14.0, 56.5, 88.9 and 85.0 % nitrate removal efficiency of thiosulfate-driven autotrophic denitrification, indicating that higher than stoichiometric S/N ratio was needed to achieve high effective thiosulfate-driven autotrophic denitrification. It is interesting that electron utilization for autotrophic denitrification in S1-S4 were approximately 26, 31, 29, and 29 %, respectively. This result indicated that thiosulfate-driven autotrophic denitrification maintained stable electron utilization efficiency for nitrate removal, even overloading thiosulfate could not promote the electron utilization.
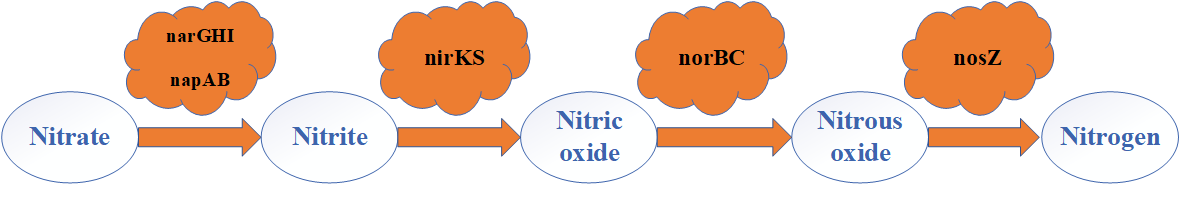
Fig.2. Nitrate reduction pathway of thiosulfate-driven autotrophic denitrification based on functional genes identification
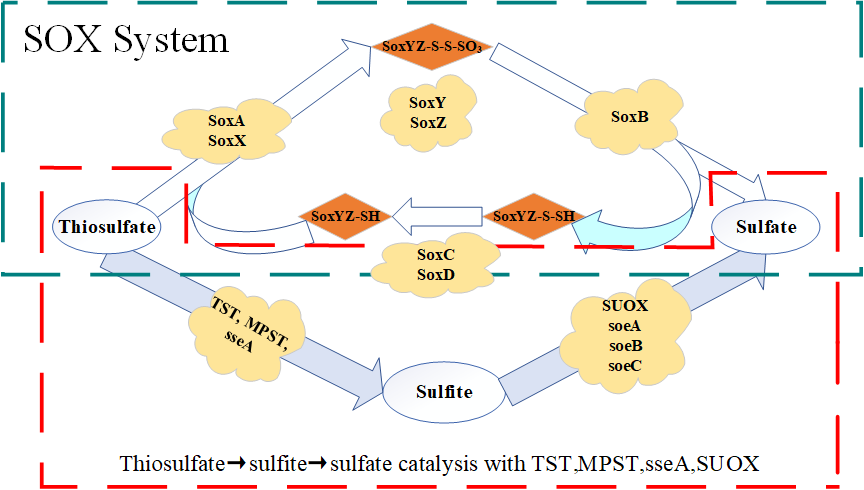
Fig.3. Thiosulfate oxidation pathway of thiosulfate-driven autotrophic denitrification based on functional genes identification
As shown in Fig. 2, the dissimilatory nitrate reduction to di-nitrogen gas is catalyzed by a series of enzymes, including nitrate reductase (nar), nitrite reductase (nir), nitric-oxide reductase (nor), and nitrous-oxide reductase (nos). Only Thiobacillus and Thauera carried a complete dissimilatory nitrate to di-nitrogen pathway. Fig.3 showed thiosulfate oxidation pathway of thiosulfate-driven autotrophic denitrification based on the metagenomes in S1-S4. None of the sludge samples taken from S1-S4 contained whole functional genes encoding for the assimilatory sulfate reduction or the dissimilatory sulfate reduction, while only the complete SOX system was identified. This result predicted that thiosulfate oxidation was only depended on the SOX system in this study, which was different from previous studies.
Furthermore, functional genes encoding for the SOX system enzymes, like SoxA, SoxX, SoxZ, and SoxY, had obvious increase during thiosulfate oxidation, responding to higher percentage of thiosulfate-driven autotrophic denitrification bacteria with thiosulfate addition (Fig.4). Additionally, SoxA and SoxX, catalyzing the first step of SOX system, had faster increased than TST, MPST, sseA, which was first step of thiosulfate → sulfite → sulfate catalysis.

Fig.4. Thiosulfate oxidation pathway of (a) All genus, (b) Thiobacillus and (c) Thauera based on functional genes identification.

Fig.5. Nitrate reduction pathway of (a) All genus, (b) Thiobacillus and (c) Thauera based on functional genes identification.
Thiobacillus and Thauera had the complete nitrate reduction pathway were shown in Fig.5. Especially, with the increase of thiosulfate, the abundance of Thiobacillus at the genus level increased, resulting in an increase in the total number of functional genes encoding nitrate reductase. As shown in Fig.4, Thiobacillus was the only genus containing full functional genes encoding for SOX system enzymes except SoxC and SoxD for catalysis from SoxYZ-S-SH to SoxYZ-S-SO3. Additionally, only Thauera was identified to carry genes encoding SoxC and SoxD in this study, meaning that Thauera might be capable of oxidizing SoxYZ-S-SH to SoxYZ-S-SO3.
Based on high abundance of Thiobacillus and Thauera in the thiosulfate- driven denitrifying microbial community, it could be plausible that Thauera would catalyze SoxYZ-S-SH, accumulated from thiosulfate reduction by Thiobacillus, into sulfate, meaning that Thiobacillus and Thauera co-worked to achieve thiosulfate reduction for electron donation to autotrophic denitrification. Thauera mainly worked for SoxYZ-S-SH catalysis and Thiobacillus completed other parts of SOX system for thiosulfate oxidation. Consequently, with thiosulfate addition, Thiobacillus showed the obvious increase with thiosulfate addition, and Thauera maintained relatively stable percentage as second dominate genus in the thiosulfate-driven autotrophic denitrification. This result indicated that parts of electrons were extracellularly transported from thiosulfate reduction, and this phenomenon caused low electron utilization for thiosulfate-driven autotrophic denitrification.
This study was funded by “Shenzhen Science and Technology Pro-gram” and “Guangdong Basic and Applied Basic Research Foundation ”.
See full text link:
https://www.sciencedirect.com/science/article/pii/S1385894723030528
