Recently, the team of Professor Lijie Zhou of the School of the College of Chemistry and Environmental Engineering, Shenzhen University published a latest research paper entitled Functional bacteria and their genes encoding for key enzymes in hydrogen-driven autotrophic denitrification with sulfate loading on the Journal of Cleaner Production (IF:11.1, JCR Chemistry Region 1, TOP Journal). The team's associate professor Lijie Zhou is the first correspondent of the paper, the master student, Fei Wu, is the first author, and Shenzhen University is the first author unit and communication unit.

Hydrogen Matrix Membrane Biofilm Reactor (H2-MBfR) is a gas transfer membrane bioreactor for autotrophic nitrogen removal. Hydrogen is considered as a high-quality electron donor for autotrophic nitrogen removal, because it will not produce harmful substances and greenhouse gases, and it is cheaper than organic electron donors. Although the water solubility of H2 is low, it can diffuse into the biofilm in a bubble-free way through the membrane pores to realize the reduction of nitrate in H2-MBfR. Sulfate not only has a great influence on the microbial community in the membrane biofilm reactor, but also has a direct influence on the functional genes of nitrate reduction, but few studies have linked them.
In order to solve the above problems, this study studied the changes of functional bacteria and their genes encoding key enzymes in the process of hydrogen autotrophic denitrification under different sulfate contents, and analyzed the degradation of nitrate and nitrite and the removal rate of nitrate. Finally, the structure and changes of microbial community were analyzed by metagenome technology, and the genes encoding key enzymes of hydrogen-driven autotrophic denitrification were analyzed to better understand the mechanism of sulfate affecting nitrate removal.
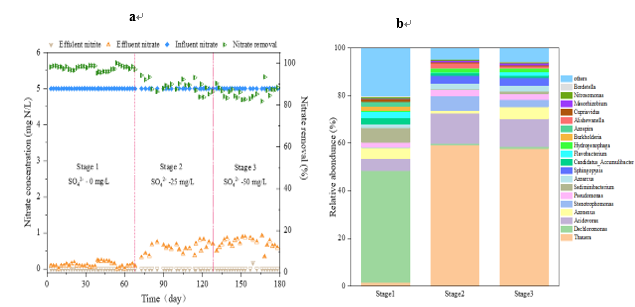
Fig 1 Performance of Hydrogen Matrix Membrane Biofilm Reactor a) Denitrification Efficiency b) Microbial Community Structure
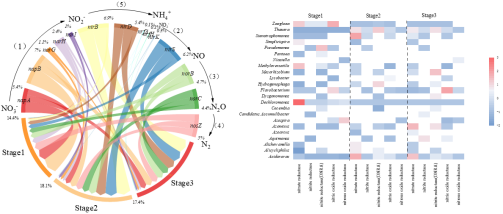
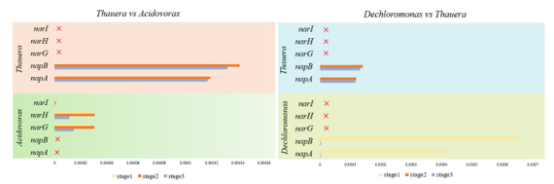
Fig. 2 Changes of functional genes at different stages in H2-MBfR
These results show that nitrate, as the only electron acceptor, is basically removed by 100%. However, SO42- led to the reduction of nitrate removal rate to below 90%. Sulfate loading replaced Dechloromonas (Stage1) and Thauera (Stages 2 and Stage 3) as dominant genera, and both of them contained all functional genes encoding denitrifying enzymes. In addition, Acidovorax is also the dominant genus (Stage 2 and Stage 3), but its abundance is far less than that of Thauera. Genes encoding nitrate reductase are the most abundant at all stages, but decrease with the increase of SO42-. Among them, napB accounts for the largest proportion of nitrate reductase functional genes in SO42-environment. Compared with napA genes, functional bacteria Dechloromonas and Thauera have more napA genes. Therefore, sulfate inhibited autotrophic denitrification, especially Dechloromonas and its functional gene napB. In order to better understand the effect of sulfate on denitrification process, especially the key microbial functional genes are of great significance for nitrate removal.
This work is supported by Guangdong Basic Sports Applied Basic Research Fund and Shenzhen Natural Science Fund.
See full text link:
https://www.sciencedirect.com/science/article/pii/S0959652624003482
